2 Fitting occupancy models with unmarked
2.1 Occupancy Modelling with Imperfect Detection
Site occupancy surveys are frequently used in the monitoring of species. Site occupancy probabilities can be used as a metric when monitoring the current state of a population. This could be for overall changes in occupancy or the expansion/contraction of species distributions. The premise being that changes in site occupancy are correlated with changes in population size and characteristics.
However, field surveys face the problem of imperfect detection; there is no guarantee that each individual, or even each species, will be detected at a site even if it is present. The detectability of a species can be defined as the probability that at least one individual is detected in a particular sampling effort, conditional to the species being present in the area of interest during sampling. To be able to estimate the effect of imperfect detection we need to perform several independent surveys of the same site.
2.1.1 The Maths
A single species, single season occupancy model like we will cover today can be seen as a hierarchical model with two processes:
- The state process: defined by the true site occupancy status (site occupied by the species or not)
- The observation process: given the occupancy status, was the species detected
The true occupancy status of a site, which is unknown, is represented by the latent variable \(z_i\), which takes the value \(1\) if site \(i\) is occupied, or \(0\) otherwise, and is modelled as:
\[z_i \sim Bernoulli(\psi_i)\]
where \(\psi_i\) is the probability of occupancy. The observation process is modelled as:
\[y_{ij} | z_i \sim Bernoulli(z_i * p_{ij})\]
where \(y_{ij}\) is our detection data site \(i\) and sampling repeat \(j\), and \(p_{ij}\) is the probability of detecting the species given it is present.
As written, these equations effectively assume the probabilities of occupancy and detection are constant across all sites, when in reality sites are heterogeneous. Covariates can be incorporated into the model using a logit-link function (like in generalised linear regression), such that the occupancy and/or detectability at site \(i\) can be modelled as a function of a series of covariates \(\{x_{i1}, ..., x_{ik}\}\).
\[logit(\psi_i) = \beta_0 + \beta_1x_{i1} + \beta_2x_{i2} ... \beta_kx_{ik}\] \[logit(p_i) = \beta_0 + \beta_1x_{i1} + \beta_2x_{i2} ... \beta_kx_{ik}\]
The four main assumptions of this model are:
- A site is closed to changes in occupancy over the survey season
- The probability of occupancy is constant across all sites, or the variation is modelled by covariates
- The probability of detection is constant across all sites and surveys, or the variation is modelled by covariates
- The detection of a species and its detection histories at each site are independent
unmarked is an R package developed by Ian Fiske and Richard Chandler (and contributed to by Marc Kéry, Andy Royle, David Miller amongst others) to help users fit models accounting for the imperfect detection of unmarked individuals.
2.2 unmarked Models
unmarked is able to fit several different models under this banner. This ranges from standard occupancy models to distance sampling and colonisation extinction models. While this guide will only cover occu() for standard occupancy models, only minimal adjustment is required to fit the others
| Model | FittingFunction | Data |
|---|---|---|
| Occupancy | occu | unmarkedFrameOccu |
| Royle-Nichols | occuRN | unmarkedFrameOccu |
| False Positives | occuFP | unmarkedFrameOccuFP |
| Point Count | pcount | unmarkedFramePCount |
| Generalised Point Count | gpcount | unmarkedFrameGPC |
| Open Point Count | pcountOpen | unmarkedFramePCO |
| Distance Sampling | distsamp | unmarkedFrameDS |
| Generalized Distance Sampling | gdistsamp | unmarkedFrameGDS |
| Arbitrary multinomial Poisson | multinomPois | unmarkedFrameMPois |
| Colinization Extinction | colext | unmarkedMultFrame |
| Generalized multinomial mixture | gmultmix | unmarkedFrameGMM |
2.4 Load Data
For this guide we will use a sample dataset supplied with the package.
| site | y.1 | y.2 | y.3 | elev | forest | length | date.1 | date.2 | date.3 | ivel.1 | ivel.2 | ivel.3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | -1.1729446 | -1.1562281 | 1.8245493 | -1.7614815 | 0.3099471 | 1.3813757 | -0.5060353 | -0.5060353 | -0.5060353 |
| 2 | 0 | 0 | 0 | -1.1265010 | -0.5014837 | 1.6292405 | -2.9043386 | -1.0471958 | 0.5956614 | -0.9336151 | -0.9907486 | -1.1621491 |
| 3 | 0 | 0 | 0 | -0.1976283 | -0.1013621 | 1.4586150 | -1.6900529 | -0.4757672 | 1.4528042 | -1.1355754 | -1.3388644 | -1.6099164 |
| 4 | 0 | 0 | 0 | -0.1047411 | 0.0077620 | 1.6863990 | -2.1900529 | -0.6900529 | 1.2385185 | -0.8193481 | -0.9272669 | -1.1970640 |
| 5 | 0 | 0 | 0 | -1.0336137 | -1.1926028 | 1.2809338 | -1.8329101 | 0.1670899 | 1.3813757 | 0.6375563 | 0.8803737 | 1.0422520 |
| 6 | 0 | 0 | 0 | -0.8478392 | 0.9171292 | 1.8082888 | -2.6186243 | 0.1670899 | 1.3813757 | -1.3288666 | -1.0422624 | -0.8989603 |
| 7 | 0 | 0 | 0 | -0.9097640 | -1.0834788 | 1.6292405 | -2.6900529 | -1.1900529 | 1.5956614 | -1.4478167 | -1.5620837 | -1.6763507 |
| 8 | 0 | 0 | 0 | -1.0026513 | -0.7924812 | 1.3083328 | -2.1186243 | -0.4757672 | 1.4528042 | -0.3205880 | -0.5568428 | -0.6355944 |
| 9 | 1 | 1 | 0 | -0.0582974 | 0.5533823 | 1.3350011 | -2.0471958 | -0.5471958 | 1.1670899 | -0.2314744 | -0.2314744 | -0.0014368 |
| 10 | 0 | 0 | 1 | -0.6311022 | 0.8080052 | 2.0412203 | -2.3329101 | -1.1186243 | -0.2614815 | -1.0968537 | -1.0211703 | -0.8319620 |
| 11 | 0 | 0 | 0 | 0.0655522 | 1.7901218 | 1.1314021 | -1.6900529 | 0.4528042 | 1.4528042 | -0.2240538 | 0.0579277 | -0.2240538 |
| 12 | 0 | 0 | 0 | -1.1110198 | -0.6469825 | 1.4816045 | -1.8329101 | -0.3329101 | 1.9528042 | 0.4168132 | 0.2843674 | 0.5492591 |
| 13 | 0 | 0 | 0 | -0.2595532 | 0.8443799 | 1.7917595 | -0.9043386 | 1.0956614 | 1.2385185 | -1.3535464 | -1.0136020 | -1.1592924 |
| 14 | 0 | 0 | 0 | -0.6465834 | -0.3196103 | 1.8245493 | -2.5471958 | -0.2614815 | 1.3099471 | -1.1169952 | -0.2240538 | -0.7880168 |
| 15 | 1 | 0 | 0 | 0.1894019 | -0.1013621 | 1.8082888 | -1.3329101 | 0.3099471 | 1.5242328 | -0.2779847 | -0.1824500 | 0.0086195 |
| 16 | 1 | 1 | 0 | 0.1429583 | 1.1353774 | 1.5040774 | -1.8329101 | -1.0471958 | 0.5242328 | -0.4826413 | -0.8840994 | -0.8193481 |
| 17 | 0 | 0 | 0 | -1.2348694 | 0.3715089 | 1.8245493 | -2.3329101 | -0.9757672 | 2.3099471 | -1.0699983 | -1.2579860 | -1.2109891 |
| 18 | 1 | 1 | 1 | -0.9716889 | -0.1377368 | 1.9315214 | -2.4043386 | -0.8329101 | 0.6670899 | -1.1149519 | -0.7348899 | -0.4815152 |
| 19 | 0 | 0 | 0 | -1.2193882 | -0.9743547 | 1.5260563 | -1.3329101 | -0.2614815 | 1.5242328 | -0.8615772 | -1.1149519 | -0.9882645 |
| 20 | 0 | 1 | 0 | 0.3132516 | 1.5718737 | 1.3083328 | 0.0242328 | 1.5242328 | 3.2385185 | -0.2418364 | -0.2418364 | -0.3993396 |
| 21 | 0 | 0 | 0 | -1.0026513 | 0.0805113 | 1.7404662 | -1.6900529 | -0.2614815 | 1.5956614 | -0.4615119 | -0.6148703 | -0.4615119 |
| 22 | 0 | 0 | 0 | 0.0191086 | -0.6106078 | 1.3862944 | -1.9757672 | -0.4757672 | 1.2385185 | -0.3579950 | -0.2123046 | 0.7346833 |
| 23 | 1 | 1 | 1 | 0.4835449 | 0.6988811 | 1.7047481 | -0.4757672 | 0.5956614 | 1.6670899 | 0.4168132 | 0.6817050 | 0.6393223 |
| 24 | 0 | 0 | 0 | -0.3988841 | -1.0471041 | 1.4586150 | -2.4043386 | -0.9757672 | 0.5242328 | -0.4579454 | -1.5421534 | -1.4337326 |
| 25 | 0 | 0 | 0 | -0.8633204 | -1.1562281 | 1.2237754 | -2.6186243 | -0.5471958 | 0.5242328 | 1.8659267 | 1.8830668 | 0.9232239 |
| 26 | 0 | 0 | 0 | -0.9871701 | -0.2832356 | 1.4350845 | -2.7614815 | -0.6186243 | 0.8813757 | -0.2643369 | 0.5681799 | -0.2643369 |
| 27 | 1 | 0 | 1 | -0.5536962 | 0.7716305 | 1.6677068 | -1.1900529 | 0.4528042 | 2.5956614 | 0.4268092 | 0.7016968 | 0.4817867 |
| 28 | 1 | 1 | 1 | 0.0655522 | 1.1353774 | 1.7227666 | -0.6186243 | 0.5242328 | 2.3099471 | -0.4204338 | -0.2643369 | -0.1082400 |
| 29 | 1 | 1 | 1 | 0.0036274 | 0.2987595 | 1.6292405 | -1.0471958 | 0.4528042 | 2.0956614 | 0.9517906 | 1.4088587 | 1.2374582 |
| 30 | 0 | 0 | 0 | -0.6930271 | 0.1896354 | 1.8245493 | -2.1900529 | -0.6186243 | 0.3099471 | 0.9038722 | 0.9132716 | 1.0542623 |
| 31 | 0 | 0 | 0 | -0.9252452 | -1.0834788 | 1.6486586 | -2.9043386 | -0.9757672 | 1.4528042 | -1.6972264 | -1.7532612 | -1.5851568 |
| 32 | 0 | 0 | 0 | -1.0645761 | -1.1562281 | 1.7047481 | -1.7614815 | -0.5471958 | -0.4043386 | -1.0665803 | -1.5433854 | -1.4374287 |
| 33 | 0 | 0 | 0 | 0.1429583 | 0.6261317 | 1.7227666 | -1.3329101 | -0.2614815 | 1.3099471 | -0.8366922 | -0.4724661 | -0.6389694 |
| 34 | 0 | 0 | 0 | -0.3834029 | 0.1168860 | 1.4350845 | -2.1186243 | -0.1186243 | 0.5956614 | 0.5681799 | 0.1519215 | 0.1519215 |
| 35 | 0 | 0 | 0 | -1.0336137 | 0.5170076 | 1.4109870 | -0.7614815 | -0.1900529 | 0.5956614 | 0.2229900 | -0.6298321 | -0.2034210 |
| 36 | 1 | 1 | NA | 1.0099061 | -0.7197319 | 1.6677068 | 1.0956614 | 2.8813757 | NA | 0.9765844 | 0.0419665 | NA |
| 37 | 1 | 1 | 1 | 1.1801994 | 0.9898786 | 1.6292405 | 0.4528042 | 1.3813757 | 2.1670899 | 0.9517906 | 1.1231912 | 1.1803247 |
| 38 | 1 | 1 | 1 | -0.1357035 | 1.7173724 | 1.2809338 | -2.1186243 | -0.2614815 | 1.5956614 | 0.3947389 | -0.0099567 | 0.1519215 |
| 39 | 0 | 0 | NA | 1.2885679 | -1.2653522 | 1.5475625 | 1.5956614 | 2.0956614 | NA | 0.3379093 | 0.5858931 | NA |
| 40 | 0 | 0 | 0 | -0.6001398 | -1.0834788 | 1.8405496 | -2.1900529 | -1.2614815 | 1.0956614 | -0.7268462 | -1.0968537 | -1.0043518 |
| 41 | 1 | 1 | 1 | 0.8550940 | 0.8443799 | 1.2809338 | -0.4757672 | 1.3099471 | 1.9528042 | -0.8193481 | -0.7384089 | -0.9812264 |
| 42 | 0 | 0 | 0 | -0.0892599 | 2.2993675 | 1.1939225 | -1.8329101 | -0.7614815 | 0.5956614 | 0.2402188 | 0.2402188 | 0.6817050 |
| 43 | 0 | 0 | 0 | -0.8633204 | -0.0649874 | 1.3862944 | -2.3329101 | -0.6186243 | 1.4528042 | -0.1394594 | -0.1394594 | 0.2976120 |
| 44 | 0 | 0 | 0 | -0.9097640 | 0.6625064 | 2.1517622 | -1.8329101 | -0.8329101 | 0.5956614 | -1.5082719 | -1.6437979 | -1.4743904 |
| 45 | 1 | 1 | 1 | 1.0253873 | -0.6469825 | 1.9315214 | 0.0956614 | 0.6670899 | 1.5956614 | -0.1014531 | -0.2281405 | -0.1014531 |
| 46 | 0 | 0 | 0 | -0.3834029 | -0.6833572 | 1.7749524 | -1.8329101 | -1.0471958 | 0.5956614 | -0.5394907 | -0.6876505 | -0.7370371 |
| 47 | 1 | 1 | 1 | -0.9871701 | 0.9171292 | 1.4586150 | -1.9043386 | -0.2614815 | 2.5956614 | 0.8973145 | 0.4907365 | 0.4907365 |
| 48 | 0 | 0 | 0 | -1.1729446 | -1.1926028 | 1.8082888 | -1.8329101 | -0.1186243 | 2.1670899 | -0.3735194 | -0.4690541 | -0.6601236 |
| 49 | 1 | 1 | 1 | 0.3442140 | 0.6261317 | 1.7404662 | -1.9757672 | -0.4043386 | 1.5956614 | -0.3081535 | -0.3081535 | -0.3081535 |
| 50 | 0 | 0 | 0 | -0.4453277 | 0.8443799 | 1.9315214 | -2.1186243 | -0.6186243 | 0.5242328 | -1.1994101 | -0.9038063 | -1.1571810 |
| 51 | 1 | 1 | 1 | 0.4371013 | 0.5533823 | 1.7749524 | -2.1900529 | -0.5471958 | 1.5242328 | -0.9839701 | -0.5888773 | -0.8358103 |
| 52 | 0 | 0 | NA | 1.0408685 | -0.8288559 | 1.5040774 | -0.5471958 | 0.8099471 | NA | -0.0423324 | -0.2365863 | NA |
| 53 | 0 | 0 | 0 | 0.3751764 | -0.2104862 | 1.3083328 | -2.6186243 | -0.8329101 | 0.5956614 | 0.0731699 | 0.5456795 | 0.2306731 |
| 54 | 0 | 0 | 0 | -0.8942828 | -0.6833572 | 1.5892352 | -1.6900529 | -1.1900529 | -0.5471958 | -1.0968537 | -1.2157847 | -1.0373882 |
| 55 | 0 | 0 | 0 | -0.9716889 | 1.7537471 | 1.1631508 | -2.3329101 | -0.2614815 | 0.5242328 | 0.3340346 | 0.4250911 | 0.7893172 |
| 56 | 0 | 0 | 0 | -0.6620647 | 1.6082484 | 1.3862944 | -2.4043386 | -1.3329101 | 0.3813757 | 0.6618381 | 0.7346833 | 0.5161476 |
| 57 | 1 | 1 | 1 | 0.9479812 | 0.5897570 | 1.7404662 | -0.4757672 | 1.0956614 | 1.6670899 | -0.2059146 | 0.1008021 | -0.0014368 |
| 58 | 0 | 0 | 0 | -0.7704331 | -0.9743547 | 1.8082888 | -2.6900529 | -0.7614815 | 0.3813757 | -1.5677034 | -1.5199360 | -1.6154707 |
| 59 | 0 | 0 | 1 | -0.2905156 | 0.3715089 | 1.4350845 | -2.9043386 | -1.0471958 | 0.0956614 | 0.5681799 | 0.5681799 | 0.5681799 |
| 60 | 0 | 0 | 0 | -1.0181325 | -1.0834788 | 1.8405496 | -2.4043386 | -1.2614815 | 0.4528042 | -1.0968537 | -1.0506028 | -1.1893556 |
| 61 | 0 | 0 | 0 | -0.6465834 | 1.0626280 | 1.5686159 | -1.9757672 | -0.9757672 | -0.4757672 | -0.3944176 | -0.3944176 | -0.2123046 |
| 62 | 0 | 0 | 0 | -1.2503507 | -0.7561065 | 2.2407097 | -2.4043386 | 0.0956614 | 1.6670899 | -1.2119890 | -1.2739849 | -1.4909707 |
| 63 | 0 | 0 | NA | 2.3722526 | -1.2653522 | 0.6418539 | 2.6670899 | 3.2385185 | NA | 0.9187133 | 0.4586383 | NA |
| 64 | 0 | 0 | 0 | -0.8168768 | -1.2653522 | 1.6292405 | -2.2614815 | -0.6186243 | 0.1670899 | -0.0194790 | -0.3622800 | -0.8193481 |
| 65 | 0 | 0 | 0 | -0.8013955 | -0.1741115 | 1.6677068 | -2.3329101 | -0.2614815 | 0.6670899 | -0.1229661 | 0.2068991 | 0.7016968 |
| 66 | 1 | 1 | 0 | -0.6930271 | -0.6469825 | 1.9315214 | -2.4043386 | -0.9757672 | 1.0242328 | -0.7348899 | -0.2281405 | -0.2281405 |
| 67 | 0 | 0 | 0 | -1.1110198 | 1.0990027 | 1.5260563 | -2.4043386 | -0.6186243 | -0.1900529 | -0.6715462 | -0.6715462 | -0.6715462 |
| 68 | 0 | 0 | 0 | -1.2658319 | -0.1741115 | 1.8718022 | -2.5471958 | -1.1900529 | 0.5956614 | -0.4756681 | -0.6101516 | -0.5204959 |
| 69 | 0 | 0 | 0 | -1.4361252 | -1.2653522 | 1.4350845 | -1.8329101 | -0.2614815 | 0.6670899 | 0.4294271 | 0.9844384 | 0.9844384 |
| 70 | 0 | 0 | NA | 1.1801994 | -1.0834788 | 1.6292405 | 1.5956614 | 2.6670899 | NA | 0.6661231 | 0.6661231 | NA |
| 71 | 0 | 0 | NA | 1.9232975 | -1.2653522 | 1.4109870 | 2.4528042 | 3.0242328 | NA | -0.4166265 | -0.9851746 | NA |
| 72 | 0 | 0 | 1 | 0.2203643 | 1.1353774 | 1.7578579 | -1.9043386 | -0.9757672 | 0.8099471 | -0.8026021 | -0.9030782 | -0.9030782 |
| 73 | 0 | 0 | 0 | -0.6001398 | -0.9743547 | 1.6863990 | -1.9757672 | -0.4757672 | 1.3099471 | -0.1718350 | -0.1718350 | 0.1519215 |
| 74 | 0 | 0 | 0 | -1.0955385 | 1.4627496 | 2.1282317 | -2.0471958 | -0.5471958 | 0.5956614 | -0.7846599 | -0.8887245 | -1.0968537 |
| 75 | 0 | 0 | 0 | -1.1884258 | 1.2081268 | 1.9600948 | -2.2614815 | -0.2614815 | 1.5242328 | -0.7919884 | -0.6688697 | -0.5457510 |
| 76 | 1 | 1 | 1 | 1.1801994 | 1.3536255 | 1.3862944 | 0.7385185 | 1.7385185 | 2.8813757 | -0.7950663 | -1.0136020 | -0.7222211 |
| 77 | 1 | 1 | 1 | 0.6073946 | 2.1902434 | 1.6677068 | -1.8329101 | -0.9043386 | 0.1670899 | 0.6467193 | 0.5917417 | 0.2068991 |
| 78 | 1 | 1 | 1 | 1.0873121 | 0.8080052 | 1.7404662 | -0.2614815 | 1.5956614 | 2.5956614 | -0.3081535 | 0.3052799 | -0.1036757 |
| 79 | 0 | 0 | 0 | 0.1739207 | 1.0626280 | 1.3862944 | -2.1900529 | 0.4528042 | 1.4528042 | 0.2976120 | 0.2976120 | 0.0062311 |
| 80 | 1 | 1 | 1 | 0.2513268 | 0.3351342 | 1.5040774 | -1.8329101 | -0.7614815 | -0.1186243 | 1.3821964 | 0.6051807 | 0.3461755 |
| 81 | 1 | 0 | 0 | -0.4608089 | 0.3351342 | 1.5686159 | -2.9043386 | -1.1900529 | -0.4757672 | -0.3944176 | -0.2123046 | -0.3944176 |
| 82 | 0 | 0 | 0 | -0.7239895 | -0.0286127 | 1.9600948 | -2.6900529 | -1.2614815 | 0.3099471 | -0.6688697 | -0.5457510 | -0.4226323 |
| 83 | 0 | 0 | 0 | -0.8478392 | -0.5014837 | 1.6863990 | -2.4043386 | -0.7614815 | 0.5242328 | 0.4756781 | 0.4756781 | 0.6375563 |
| 84 | 0 | 0 | 0 | -1.1265010 | -0.4287343 | 1.5686159 | -1.8329101 | -0.8329101 | 0.4528042 | -0.9407568 | -0.9407568 | -0.9407568 |
| 85 | 0 | NA | NA | 1.8304102 | -1.2289775 | 2.1860513 | 0.4528042 | 1.5956614 | NA | -0.6010852 | NA | NA |
| 86 | 1 | 1 | 1 | 0.7312443 | 0.2987595 | 1.6094379 | -1.9043386 | -0.8329101 | 0.5956614 | 0.7346833 | 0.5598548 | 0.0353692 |
| 87 | 1 | 1 | 1 | 1.3504927 | -0.1741115 | 1.4109870 | 0.5242328 | 1.3099471 | 1.6670899 | 1.0758121 | 0.8626066 | 1.0758121 |
| 88 | 0 | 0 | 0 | -0.6001398 | -0.3196103 | 1.5686159 | -2.4043386 | -0.9757672 | 0.5956614 | -0.2123046 | 0.3340346 | -0.0301915 |
| 89 | 0 | NA | NA | -0.6930271 | -1.0107294 | 1.7227666 | -2.9043386 | -0.9043386 | 1.0956614 | -1.5131121 | NA | NA |
| 90 | 0 | 0 | 0 | -1.0645761 | 1.8992459 | 1.8405496 | -2.7614815 | -1.4043386 | 0.0242328 | -0.9581009 | -1.1431046 | -0.2643369 |
| 91 | 0 | 0 | 0 | -0.5227338 | 0.5170076 | 1.5686159 | -2.3329101 | -0.5471958 | 1.3813757 | 0.5161476 | 0.7589650 | 0.8196694 |
| 92 | 0 | 0 | 0 | -1.1265010 | 0.1532607 | 2.0281482 | -2.1900529 | -0.5471958 | 1.0956614 | -0.6915494 | -0.6148703 | -0.6148703 |
| 93 | 1 | 1 | 1 | -0.2595532 | 2.1174940 | 1.6863990 | -2.4043386 | -1.1900529 | -0.2614815 | -0.1718350 | 0.3677592 | -0.0639162 |
| 94 | 0 | 0 | 0 | 0.4216201 | 0.4806329 | 1.5475625 | -0.1900529 | 1.1670899 | 1.6670899 | 0.0899256 | 0.2139175 | 0.2139175 |
| 95 | 0 | 0 | 0 | -0.5072525 | -0.8652306 | 1.1939225 | -2.2614815 | -1.6900529 | 0.3813757 | -0.7310508 | -0.4661591 | -0.5544564 |
| 96 | 1 | 0 | 1 | 0.0965147 | 1.3900002 | 1.8245493 | -2.2614815 | -0.8329101 | 0.1670899 | 0.4809000 | 0.1989185 | 0.6218907 |
| 97 | 0 | 0 | 0 | -0.9407264 | -0.8288559 | 1.7749524 | -1.9043386 | -0.9043386 | 1.0956614 | -1.4284494 | -1.1321298 | -1.2802896 |
| 98 | 1 | 1 | 1 | 0.4990261 | 0.2623848 | 1.7227666 | -0.2614815 | 1.0956614 | 1.7385185 | -0.0041754 | 0.2559861 | 0.6722445 |
| 99 | 0 | 0 | 0 | 0.3751764 | -0.3196103 | 1.5892352 | -0.9757672 | -0.4757672 | 0.7385185 | -0.4427333 | -0.2643369 | 0.2708525 |
| 100 | 0 | 0 | 0 | -0.6156210 | -0.7924812 | 1.1631508 | -2.1900529 | -1.1900529 | 0.5242328 | -0.3944176 | 0.0608650 | -0.3944176 |
| 101 | 0 | 0 | 0 | -1.0955385 | -1.1926028 | 1.5040774 | -2.4043386 | -0.9043386 | -0.1186243 | -0.3660889 | -1.1431046 | -1.2726072 |
| 102 | 0 | 0 | NA | 1.3969363 | -0.7197319 | 1.4816045 | 1.3099471 | 3.2385185 | NA | -0.3116390 | -0.0467473 | NA |
| 103 | 1 | 1 | 1 | 1.3040491 | 0.6988811 | 1.7578579 | -0.7614815 | 1.3813757 | 3.1670899 | 0.1519215 | 0.1519215 | -0.1997451 |
| 104 | 1 | 1 | 1 | 0.8860564 | 1.4263749 | 1.7047481 | -0.2614815 | 0.3813757 | 1.5956614 | 0.4168132 | 0.7346833 | -0.0070135 |
| 105 | 1 | 1 | 1 | 0.7002819 | 0.3715089 | 1.4586150 | -0.4043386 | 0.4528042 | 1.5242328 | 0.5584995 | 0.9650775 | 0.5584995 |
| 106 | 0 | 0 | 0 | -0.8323580 | 0.9535039 | 1.7749524 | -2.3329101 | -0.8329101 | 0.6670899 | -0.1937846 | -0.2925578 | -0.1443980 |
| 107 | 0 | 0 | 0 | -1.0026513 | -0.0649874 | 1.7047481 | -2.5471958 | -1.6186243 | 0.5242328 | -0.4308402 | -0.1659485 | -0.2189269 |
| 108 | 0 | 0 | 0 | -0.8013955 | 0.6988811 | 1.2527630 | -1.9757672 | -0.5471958 | 0.5956614 | -0.3475886 | 0.0353692 | -0.0978335 |
| 109 | 1 | 1 | 1 | 0.2822892 | 1.2445015 | 1.2527630 | -0.8329101 | 0.5242328 | 1.6670899 | 0.9844384 | 0.9844384 | 0.9844384 |
| 110 | 0 | 0 | 0 | 0.1739207 | 0.4442583 | 1.9459101 | -1.6186243 | -1.1186243 | 1.5956614 | -1.0136020 | -1.0552278 | -1.0136020 |
| 111 | 0 | 0 | 0 | -0.7859143 | -1.0834788 | 1.8718022 | -1.1900529 | -0.9757672 | 0.0956614 | -1.1480855 | -1.0584298 | -1.1480855 |
| 112 | 0 | 0 | 0 | -1.0800573 | 0.0441367 | 2.0412203 | -2.3329101 | -0.2614815 | 1.0956614 | -0.6049119 | -0.2643369 | -0.4913869 |
| 113 | 1 | 1 | 1 | 1.2730867 | 0.9171292 | 1.5686159 | 1.2385185 | 1.5956614 | 1.9528042 | 0.8803737 | 1.2445999 | 1.2445999 |
| 114 | 0 | 1 | 1 | -0.5846586 | 1.3536255 | 1.1939225 | -1.9757672 | -0.1900529 | 0.5242328 | -0.5544564 | -0.0246729 | 0.2402188 |
| 115 | 0 | 0 | 0 | -0.2595532 | -0.2104862 | 1.5260563 | -2.3329101 | -0.5471958 | 1.4528042 | -0.3548278 | -0.4815152 | -0.6082025 |
| 116 | 0 | 0 | 0 | -0.7704331 | -1.0107294 | 1.7227666 | -1.9757672 | -0.5471958 | 0.5956614 | -0.5244984 | -0.4724661 | -0.8366922 |
| 117 | 0 | 0 | 0 | -1.0645761 | -1.0107294 | 1.4586150 | -2.3329101 | -0.9043386 | 0.6670899 | 0.3552105 | 0.6940255 | 1.0328405 |
| 118 | 0 | 0 | 0 | -1.2039070 | -0.7924812 | 1.4586150 | -2.3329101 | -0.9757672 | 0.6670899 | -0.7289974 | -0.7289974 | -0.3224194 |
| 119 | 0 | 0 | 0 | -1.3277567 | -1.1926028 | 2.1162555 | -2.3329101 | -1.0471958 | 0.5956614 | -1.3927482 | -1.4980666 | -1.2172176 |
| 120 | 1 | 1 | 1 | 0.3442140 | -0.6833572 | 1.6863990 | -0.3329101 | 2.6670899 | 2.7385185 | -0.0099567 | -0.0099567 | 0.1519215 |
| 121 | 0 | 0 | 0 | -0.9097640 | 0.9898786 | 1.3862944 | -2.3329101 | -0.8329101 | 0.5956614 | 0.0062311 | 0.4433024 | 0.3704572 |
| 122 | 0 | 0 | 0 | -1.1574634 | -0.9379800 | 1.9600948 | -1.8329101 | -0.2614815 | 0.6670899 | -0.9971862 | -0.5047114 | -0.4226323 |
| 123 | 0 | 0 | 0 | -1.1884258 | 0.0805113 | 2.0014800 | -2.3329101 | -0.4043386 | 0.6670899 | -1.1081039 | -0.8718491 | -0.9899765 |
| 124 | 0 | 0 | 0 | -1.0336137 | 1.3172508 | 1.7404662 | -2.5471958 | -0.5471958 | 0.1670899 | -0.5126314 | -0.6659897 | -0.6148703 |
| 125 | 1 | 1 | 1 | 1.1647182 | 0.2260101 | 1.5686159 | 2.1670899 | 3.0956614 | 3.5242328 | 1.9730521 | 1.7909390 | 1.6088260 |
| 126 | 1 | 0 | NA | 0.5919134 | -0.5742331 | 1.0986123 | 1.8813757 | 3.0242328 | NA | 3.0657304 | 3.0657304 | NA |
| 127 | 0 | 0 | 0 | -0.7394707 | -0.3923596 | 1.9021075 | -2.5471958 | -1.1900529 | 0.8813757 | -0.4134444 | -0.2829753 | 0.1084318 |
| 128 | 0 | 0 | 0 | -1.1419822 | -0.9016053 | 1.8718022 | -2.6900529 | -0.5471958 | -0.2614815 | -0.8342907 | -0.6549794 | -0.8522218 |
| 129 | 0 | 0 | 0 | -1.0800573 | -1.2653522 | 1.8082888 | -1.9757672 | 0.3099471 | 0.8099471 | -1.3766339 | -1.3766339 | -1.4244013 |
| 130 | 0 | 0 | 0 | -1.0645761 | 1.2808761 | 1.8245493 | -2.6900529 | -0.8329101 | 0.8813757 | -0.6470261 | 0.0579277 | 0.2459154 |
| 131 | 1 | 1 | 1 | 1.0408685 | 0.3715089 | 1.7749524 | 1.0956614 | 1.5956614 | 2.0956614 | -0.9839701 | -0.5394907 | -0.5394907 |
| 132 | 0 | 0 | 0 | -1.0645761 | -0.6106078 | 1.4350845 | -2.7614815 | -1.3329101 | 1.3813757 | -0.6805953 | -1.0274773 | -0.8193481 |
| 133 | 0 | 0 | 0 | -1.0800573 | -0.5378584 | 1.8718022 | -2.7614815 | -1.0471958 | 0.5242328 | -1.1480855 | -1.1480855 | -1.1480855 |
| 134 | 0 | 0 | 0 | -1.1419822 | -1.1562281 | 1.6094379 | -2.4043386 | -0.9043386 | 1.0956614 | -1.0136020 | -0.6639449 | -0.6639449 |
| 135 | 0 | 0 | 0 | -0.5536962 | -0.5378584 | 1.8082888 | -1.9757672 | -0.6186243 | 1.3099471 | 0.1041542 | 0.1041542 | 0.5818278 |
| 136 | 0 | 0 | 0 | -0.7859143 | 0.0441367 | 1.4586150 | -2.2614815 | -1.1900529 | 0.5242328 | 1.1683665 | 1.5071815 | 1.1006035 |
| 137 | 0 | 0 | 0 | -0.8788016 | -0.5742331 | 1.6863990 | -2.5471958 | -0.5471958 | 0.7385185 | -1.1215209 | -0.3337133 | -0.1610431 |
| 138 | 1 | 0 | 0 | 0.8086503 | 0.0077620 | 1.2809338 | 0.5956614 | 1.5242328 | 1.6670899 | 0.5566172 | 0.3137998 | 0.8803737 |
| 139 | 0 | 0 | 0 | -0.4298465 | 1.8992459 | 1.9459101 | -2.6900529 | -0.1900529 | 0.8099471 | -1.0968537 | -0.8887245 | -0.8470986 |
| 140 | 1 | 1 | 1 | 0.1119959 | 0.5897570 | 1.7578579 | -0.9043386 | -0.4757672 | 0.5242328 | -0.6518878 | -0.6518878 | -0.6518878 |
| 141 | 0 | 0 | 0 | -0.4143653 | -0.5014837 | 1.4350845 | -1.1186243 | -0.1186243 | 1.5242328 | 0.5681799 | 0.9844384 | 2.6494720 |
| 142 | 0 | 0 | 0 | -0.9871701 | -0.8652306 | 1.8082888 | -2.4043386 | -0.1900529 | 2.0242328 | -0.4690541 | -0.1824500 | -0.1824500 |
| 143 | 0 | 0 | 0 | -1.1265010 | -0.2104862 | 1.5475625 | -2.3329101 | -0.6186243 | 0.8813757 | -1.0879971 | -0.7160215 | -0.9020093 |
| 144 | 0 | 0 | 0 | -1.0955385 | 0.7352558 | 1.5892352 | -2.8329101 | -0.9757672 | 0.7385185 | -0.0859404 | -0.0264749 | 0.5087145 |
| 145 | 0 | 1 | NA | 1.0873121 | -0.1013621 | 1.9315214 | 0.7385185 | 2.1670899 | NA | -0.7348899 | -0.4815152 | NA |
| 146 | 1 | 0 | 1 | 0.3442140 | -0.1013621 | 1.5040774 | -1.0471958 | -0.4757672 | 1.0956614 | -0.0423324 | -0.0423324 | 0.1519215 |
| 147 | 0 | 0 | 0 | -1.0955385 | -1.1562281 | 1.6486586 | -1.9757672 | -0.5471958 | 0.5956614 | -1.2489481 | -1.4170525 | -1.3610177 |
| 148 | 0 | 0 | NA | -0.5382150 | 0.8443799 | 1.6094379 | -0.7614815 | 0.0956614 | 1.3099471 | -0.3725641 | -0.3142879 | NA |
| 149 | 0 | 1 | 1 | 1.1956806 | 0.0441367 | 1.4816045 | 1.1670899 | 2.1670899 | 3.1670899 | 1.0128196 | 1.1452655 | 1.2777113 |
| 150 | 0 | 0 | 0 | 0.7467255 | 1.8628712 | 1.6094379 | -0.1900529 | 1.3099471 | 2.1670899 | -0.0811832 | -0.3725641 | -0.4424955 |
| 151 | 0 | 1 | 1 | 0.0500710 | 1.4627496 | 1.5260563 | -2.6900529 | -0.5471958 | 1.4528042 | 0.3419525 | 0.2152652 | 0.6586709 |
| 152 | 1 | 0 | 0 | -1.2348694 | -0.4287343 | 1.7578579 | -1.9043386 | -0.6186243 | 0.5956614 | -0.4509355 | -0.5514116 | -0.5514116 |
| 153 | 0 | 0 | NA | 1.0253873 | -1.2653522 | 1.3350011 | 0.7385185 | 2.4528042 | NA | 1.2254301 | 1.2254301 | NA |
| 154 | 0 | 0 | 0 | -0.3524404 | 1.3172508 | 1.9315214 | -2.5471958 | -0.2614815 | 2.1670899 | -0.8615772 | -0.8615772 | -1.1149519 |
| 155 | 0 | 0 | 0 | -0.4608089 | 1.2445015 | 1.9021075 | -2.3329101 | -1.0471958 | 0.6670899 | -1.1962587 | -0.9353206 | -1.1527690 |
| 156 | 0 | 0 | 0 | -1.0800573 | -1.0471041 | 1.5260563 | -1.8329101 | -0.4757672 | 0.6670899 | -0.4815152 | -0.1014531 | -0.1014531 |
| 157 | 0 | 0 | 0 | -0.9716889 | 1.1717521 | 1.5892352 | -1.7614815 | -1.1186243 | -0.2614815 | 0.8060419 | 0.8060419 | 0.8060419 |
| 158 | 0 | 0 | 0 | -0.9716889 | 1.1717521 | 1.5892352 | -1.7614815 | -1.1186243 | -0.2614815 | 0.8060419 | 0.8060419 | 0.8060419 |
| 159 | 0 | 0 | 0 | -1.2967943 | 0.2623848 | 1.6094379 | -2.4043386 | -0.8329101 | 1.5956614 | 0.7346833 | 0.8512357 | 1.2008927 |
| 160 | 0 | 0 | 0 | -0.9716889 | 1.3536255 | 1.7047481 | -2.4043386 | -0.8329101 | 1.1670899 | -0.0599918 | -0.0599918 | 0.0989432 |
| 161 | 0 | 0 | 0 | 0.2203643 | 1.6082484 | 1.3083328 | -0.5471958 | 1.2385185 | 1.7385185 | 0.4669279 | 0.0731699 | -0.0843332 |
| 162 | 0 | 0 | 0 | -1.4206440 | 0.4442583 | 1.9878743 | -2.7614815 | -1.7614815 | -0.1186243 | -1.4846012 | -1.2451101 | -1.3249405 |
| 163 | 1 | 0 | 0 | 1.2885679 | -0.3196103 | 1.4109870 | 0.9528042 | 2.3813757 | 3.8099471 | 1.5022232 | 1.5022232 | 1.0758121 |
| 164 | 0 | 0 | 0 | -0.0737787 | 0.7716305 | 1.5892352 | -2.3329101 | -0.4043386 | 0.5956614 | -0.2643369 | -0.2643369 | -0.3832678 |
| 165 | 1 | 0 | NA | 1.1337558 | -0.6106078 | 1.4816045 | -0.4043386 | 1.5956614 | NA | 0.6154820 | 0.6154820 | NA |
| 166 | 1 | 1 | 1 | 0.7002819 | -0.0649874 | 1.0647107 | 0.0956614 | 1.5956614 | 3.3813757 | 3.0657304 | 3.2666827 | 2.9652542 |
| 167 | 0 | 0 | 0 | 0.5609510 | -0.5014837 | 1.1939225 | -0.4757672 | 0.5956614 | 1.7385185 | -0.1129702 | -0.3778619 | -0.5544564 |
| 168 | 1 | 1 | 1 | 0.2513268 | 0.1532607 | 1.3862944 | -0.5471958 | -0.2614815 | 0.5956614 | 2.1187425 | 1.6088260 | 1.6088260 |
| 169 | 0 | 0 | 0 | 0.0036274 | 1.2445015 | 1.7047481 | -1.6900529 | -0.6186243 | 0.3813757 | 0.0989432 | 0.0989432 | 0.2578782 |
| 170 | 0 | 0 | 0 | -0.5382150 | 0.2623848 | 1.7917595 | -2.3329101 | 0.1670899 | 1.3099471 | 0.1519215 | 0.2976120 | 0.1033581 |
| 171 | 0 | 0 | 0 | -0.9871701 | 2.2993675 | 1.7578579 | -1.6186243 | -0.2614815 | 0.5956614 | -0.3504593 | -0.6518878 | -0.6016497 |
| 172 | 1 | 1 | 1 | 0.3596952 | 0.6261317 | 1.2527630 | -2.1186243 | -0.5471958 | 0.4528042 | 0.3184249 | 0.3184249 | 0.7346833 |
| 173 | 0 | 1 | 0 | 0.2203643 | 2.0083700 | 1.4816045 | -2.1900529 | -0.6186243 | -0.2614815 | -0.5103078 | 0.0856986 | -0.1791931 |
| 174 | 1 | 0 | 1 | -0.4453277 | 0.5533823 | 1.4109870 | -2.2614815 | -0.4757672 | 1.4528042 | 0.4361956 | 0.0097845 | 0.4361956 |
| 175 | 0 | 0 | 0 | -0.9252452 | 0.6988811 | 1.5892352 | -2.6900529 | -1.6900529 | -0.6186243 | -0.0859404 | -0.2643369 | -0.4427333 |
| 176 | 0 | 0 | 0 | -1.0490949 | -0.9379800 | 1.6292405 | -1.6186243 | -1.0471958 | -0.8329101 | -0.3622800 | -0.3622800 | -0.3622800 |
| 177 | 1 | 1 | 1 | 0.0655522 | 0.5897570 | 1.6094379 | -1.9043386 | 0.0956614 | 1.8813757 | 0.5015786 | 0.3850262 | 0.3850262 |
| 178 | 1 | 0 | 0 | -0.7239895 | 0.0077620 | 1.6486586 | -2.9043386 | -0.2614815 | 0.4528042 | 0.7683042 | 0.6001998 | 1.1045129 |
| 179 | 0 | 0 | 0 | 0.9944249 | 0.8080052 | 1.3350011 | 0.5242328 | 2.1670899 | 2.5956614 | 2.7590137 | 1.3787884 | 1.8388635 |
| 180 | 0 | 0 | 0 | -1.0181325 | -0.6106078 | 1.7578579 | -2.3329101 | -1.0471958 | -0.5471958 | -0.4509355 | -0.1997451 | -0.0992689 |
| 181 | 1 | 1 | NA | 1.1027933 | -0.8288559 | 1.5260563 | 1.5956614 | 3.0242328 | NA | 1.1020766 | 0.9120456 | NA |
| 182 | 0 | 0 | 0 | 0.7312443 | -0.0286127 | 1.2237754 | -0.6186243 | 0.6670899 | 2.0956614 | 0.0662213 | -0.0194790 | -0.4479803 |
| 183 | 0 | 0 | 0 | 0.3287328 | 1.2081268 | 1.4109870 | -1.0471958 | -0.2614815 | 0.4528042 | 1.2890177 | 1.2890177 | 1.5022232 |
| 184 | 0 | 0 | 0 | 0.3287328 | 1.0262533 | 1.5475625 | -0.9757672 | 1.3813757 | 2.5956614 | 0.0279297 | -0.9020093 | -0.2820500 |
| 185 | 1 | 0 | 0 | 0.1739207 | 0.4806329 | 1.5040774 | -1.8329101 | -0.6186243 | 0.0956614 | 0.9936885 | 1.1231912 | 1.1231912 |
| 186 | 0 | 1 | 1 | 0.9789437 | -0.0649874 | 1.2527630 | -0.4757672 | 0.5242328 | 3.3813757 | 1.7337035 | 1.2341934 | 1.2341934 |
| 187 | 1 | 1 | 0 | -0.0737787 | 2.0083700 | 1.6094379 | -2.9043386 | -1.0471958 | 0.0956614 | 0.4433024 | 0.2684739 | 0.5598548 |
| 188 | 0 | 0 | 0 | -1.0800573 | -0.0649874 | 1.7917595 | -2.9043386 | -1.6186243 | -0.4043386 | -0.1394594 | 0.1519215 | -0.4308402 |
| 189 | 0 | 0 | 0 | -0.2131096 | 0.0805113 | 1.3862944 | -1.8329101 | -0.4043386 | 0.6670899 | 0.7346833 | 0.7346833 | 0.5161476 |
| 190 | 0 | 0 | 0 | -1.1884258 | -0.9743547 | 1.2237754 | -1.8329101 | -0.2614815 | 1.9528042 | -0.4479803 | 0.3233221 | -0.4479803 |
| 191 | 1 | 1 | 1 | 1.3195303 | 0.0441367 | 1.5475625 | -0.0471958 | 1.6670899 | 2.9528042 | 0.3999053 | 1.2678483 | 1.1438565 |
| 192 | 1 | 1 | 1 | 1.0873121 | 1.1353774 | 1.5260563 | -0.6900529 | 0.8813757 | 1.7385185 | 0.6586709 | 0.5319836 | 0.2786089 |
| 193 | 0 | 0 | 0 | -0.3524404 | 0.7716305 | 1.4109870 | -1.6186243 | -1.0471958 | -0.4757672 | 1.1468807 | 0.5072641 | 1.2890177 |
| 194 | 0 | 0 | 0 | -1.1265010 | -1.2653522 | 1.7749524 | -2.1900529 | -1.1900529 | 0.8099471 | -0.1937846 | -0.0950114 | 0.0531484 |
| 195 | 0 | 0 | 0 | -0.3524404 | 0.2260101 | 1.6094379 | -2.9043386 | -0.5471958 | 0.5956614 | 0.2101977 | 0.3850262 | 0.7346833 |
| 196 | 0 | 0 | 0 | -0.1666659 | 1.0262533 | 2.2082744 | -2.1186243 | 0.0242328 | 1.7385185 | -0.7446350 | -0.4884760 | -0.3283766 |
| 197 | 0 | 0 | NA | 1.2421242 | -1.2653522 | 1.7047481 | 1.7385185 | 2.3099471 | NA | -0.2719052 | -0.8546670 | NA |
| 198 | 0 | 1 | 0 | 0.2668080 | 0.0077620 | 1.5475625 | -1.8329101 | 0.0956614 | 1.5956614 | -0.3440459 | -0.2820500 | -0.2820500 |
| 199 | 0 | 0 | 0 | -0.9871701 | -0.7561065 | 2.0149030 | -1.8329101 | -0.1900529 | 0.6670899 | -0.8970497 | -1.2078559 | -0.9359004 |
| 200 | 0 | 0 | 0 | -1.1729446 | -1.2289775 | 1.9740810 | -2.1900529 | -0.9043386 | 0.5242328 | -0.8193481 | -1.3049829 | -1.0621655 |
| 201 | 1 | 1 | 0 | 0.3751764 | -0.2832356 | 1.3609766 | -1.1186243 | -0.1186243 | 1.3099471 | 1.4220433 | 1.4967564 | 1.5565268 |
| 202 | 0 | 0 | NA | 1.6446357 | -1.2653522 | 1.8870696 | 0.6670899 | 1.6670899 | NA | -0.6427536 | -0.9076453 | NA |
| 203 | 1 | 0 | NA | 1.2421242 | -1.0471041 | 1.4109870 | 0.6670899 | 1.8813757 | NA | 2.3550453 | 2.3550453 | NA |
| 204 | 0 | 1 | NA | 1.7839666 | -1.2653522 | 1.3350011 | 0.6670899 | 1.5956614 | NA | 1.3787884 | 0.9187133 | NA |
| 205 | 0 | 0 | NA | 1.3504927 | -1.2289775 | 1.3609766 | 0.5956614 | 1.0956614 | NA | -0.0722176 | -0.0722176 | NA |
| 206 | 1 | 1 | 1 | 0.0036274 | 0.8443799 | 1.4350845 | -1.9757672 | -0.9043386 | 0.3813757 | 1.1925676 | 1.2619440 | 1.3313204 |
| 207 | 1 | 0 | NA | 1.2885679 | -0.9379800 | 1.4816045 | 1.5956614 | 2.5956614 | NA | 1.6088260 | 1.3439342 | NA |
| 208 | 1 | 1 | NA | 1.6136733 | -0.7561065 | 1.5260563 | 1.6670899 | 2.2385185 | NA | -0.2914842 | 0.0252342 | NA |
| 209 | 1 | 1 | NA | 1.7530042 | -1.1926028 | 1.4586150 | 2.3813757 | 2.8813757 | NA | 1.9815224 | 2.1848114 | NA |
| 210 | 1 | 1 | 1 | 1.5672297 | -0.1741115 | 1.9169226 | 0.6670899 | 1.4528042 | 2.4528042 | 0.3233221 | 0.3233221 | 0.3233221 |
| 211 | 1 | 1 | 1 | 0.9170188 | 2.1174940 | 2.1400662 | -0.1900529 | 0.5956614 | 1.3099471 | -1.3564030 | -1.0478821 | -1.5278036 |
| 212 | 1 | 1 | 1 | 1.0099061 | 1.7173724 | 1.8718022 | 0.4528042 | 1.3099471 | 1.4528042 | 0.1070937 | -0.1170454 | 0.0174380 |
| 213 | 1 | 1 | 1 | 1.0718309 | 1.6809977 | 1.8245493 | 0.3813757 | 0.5242328 | 1.3813757 | -0.0360661 | 0.0579277 | 0.1049246 |
| 214 | 1 | 1 | 1 | 1.0253873 | 1.7173724 | 1.5260563 | 0.0956614 | 1.0242328 | 2.0242328 | 1.0387329 | 1.0387329 | 1.0387329 |
| 215 | 0 | 0 | NA | 1.9232975 | -1.2653522 | 1.6677068 | 1.4528042 | 2.4528042 | NA | 0.4927822 | 0.4268092 | NA |
| 216 | 1 | 1 | 1 | 0.9944249 | 1.2445015 | 1.5475625 | -0.2614815 | 0.8813757 | 1.5956614 | 1.2802475 | 0.9082719 | 0.7470825 |
| 217 | 1 | 1 | 1 | 1.0408685 | 0.6625064 | 1.7749524 | 0.8099471 | 1.4528042 | 2.9528042 | 0.2013081 | 0.2013081 | 0.4976277 |
| 218 | 0 | 0 | NA | 1.6601169 | -1.2653522 | 1.2809338 | 2.0956614 | 3.2385185 | NA | 2.0944608 | 2.0944608 | NA |
| 219 | 0 | 0 | NA | 1.6601169 | -1.2653522 | 1.4109870 | 0.4528042 | 2.8813757 | NA | -0.2034210 | -0.2034210 | NA |
| 220 | 0 | 0 | NA | 1.9697411 | -1.2653522 | 1.4586150 | 1.5956614 | 2.1670899 | NA | 1.7104705 | 1.7104705 | NA |
| 221 | 0 | 0 | NA | 1.8149290 | -1.2653522 | 2.2300144 | 0.1670899 | 2.0956614 | NA | -0.5060353 | -0.8820106 | NA |
| 222 | 0 | 0 | NA | 2.2793653 | -1.2653522 | 0.6931472 | 2.3813757 | 3.4528042 | NA | 0.1519215 | 0.0062311 | NA |
| 223 | 0 | 0 | NA | 2.4341774 | -1.2653522 | 1.6486586 | 2.3099471 | 2.5242328 | NA | 1.3286520 | 1.1605477 | NA |
| 224 | 0 | 0 | NA | 1.9697411 | -1.2653522 | 1.7749524 | 2.0956614 | 3.0956614 | NA | 0.2506947 | 0.5470143 | NA |
| 225 | 0 | 0 | NA | 1.1956806 | -1.2653522 | 1.6486586 | 0.5956614 | 1.4528042 | NA | -0.9127394 | -0.5765307 | NA |
| 226 | 0 | 0 | NA | 1.5053048 | -1.2653522 | 1.5892352 | 1.8099471 | 2.1670899 | NA | 0.0924560 | 0.0924560 | NA |
| 227 | 0 | 0 | NA | 1.0408685 | -1.2289775 | 1.2527630 | 1.5242328 | 1.7385185 | NA | 0.7346833 | -0.3475886 | NA |
| 228 | 0 | 0 | NA | 1.3504927 | -1.2653522 | 1.7404662 | 2.3813757 | 3.2385185 | NA | -1.2283037 | -0.6148703 | NA |
| 229 | 0 | 0 | NA | 1.0408685 | -1.2653522 | 0.3364722 | 1.3813757 | 1.8099471 | NA | 5.3551516 | 5.9795392 | NA |
| 230 | 0 | 0 | NA | 1.3504927 | -0.7197319 | 1.2237754 | 2.2385185 | 2.6670899 | NA | 0.0662213 | 0.3233221 | NA |
| 231 | 0 | 0 | NA | 1.5053048 | -1.2653522 | 0.1823216 | 2.0956614 | 2.6670899 | NA | 5.4939044 | 5.4939044 | NA |
| 232 | 0 | 0 | NA | 1.3504927 | -1.2653522 | 1.4816045 | 1.1670899 | 2.1670899 | NA | -0.3778619 | 0.6154820 | NA |
| 233 | 0 | 0 | NA | 1.1956806 | -1.1926028 | 1.6677068 | 0.6670899 | 1.5956614 | NA | -0.6177638 | -0.4528312 | NA |
| 234 | 0 | 0 | NA | 1.1956806 | -1.2653522 | 1.6486586 | 1.1670899 | 3.1670899 | NA | 0.2639911 | 0.2639911 | NA |
| 235 | 0 | 0 | NA | 1.6601169 | -1.2653522 | 1.4350845 | 0.6670899 | 2.6670899 | NA | -0.1255841 | 0.1519215 | NA |
| 236 | 0 | 0 | NA | 1.6601169 | -1.2653522 | 0.9932518 | 1.7385185 | 2.8099471 | NA | 3.7132435 | 3.7132435 | NA |
| 237 | 0 | 0 | NA | 1.8149290 | -1.2653522 | 1.0647107 | 1.8099471 | 3.3099471 | NA | 1.0562070 | 2.2619210 | NA |
Data needs to be formatted for use with a specific model fitting function, and this is accomplished with a call to an appropriate unmarkedFrame. unmarkedFrame’s are a special class of object used in this package for organising our data. These are built using R’s S4 reference class system instead of the more common S3 (like a linear model object lm). The benefit of this is a formal definition and validity checking to prevent accidentally breaking it, but the consequence is a slightly different syntax for examining it (covered later). In our case, to fit an occu() model we need to use the unmarkedFrameOccu(). This requires two data.frames and a list of data.frames:
y: Adata.frameof presence-absence records. Rows are sites, columns are repeat visits.siteCovs: Adata.frameof the site-level covariates. These are things that don’t change between visits like elevation, annual rainfall, distance from roads, etc.. One column per covariateobsCovs: Alistofdata.frames for the observation-level covariates. Each covariate is its owndata.framewith rows as sites and columns as repeat visits. These are things that can change between visits. Could be environmental conditions like daily temperature or cloud cover, or methodological variables like survey method (spotlighting, pitfall traps, ink cards etc.) or an observer ID.
y <- data[ ,2:4]
siteCovs <- data[ ,5:7]
obsCovs <- list(date = data[ ,8:10],
ivel = data[ ,11:13])
umf <- unmarkedFrameOccu(y = y, siteCovs = siteCovs, obsCovs = obsCovs)
summary(umf)## unmarkedFrame Object
##
## 237 sites
## Maximum number of observations per site: 3
## Mean number of observations per site: 2.81
## Sites with at least one detection: 79
##
## Tabulation of y observations:
## 0 1 <NA>
## 483 182 46
##
## Site-level covariates:
## elev forest
## Min. :-1.436125 Min. :-1.265352
## 1st Qu.:-0.940726 1st Qu.:-0.974355
## Median :-0.166666 Median :-0.064987
## Mean : 0.007612 Mean : 0.000088
## 3rd Qu.: 0.994425 3rd Qu.: 0.808005
## Max. : 2.434177 Max. : 2.299367
## length
## Min. :0.1823
## 1st Qu.:1.4351
## Median :1.6094
## Mean :1.5924
## 3rd Qu.:1.7750
## Max. :2.2407
##
## Observation-level covariates:
## date ivel
## Min. :-2.90434 Min. :-1.7533
## 1st Qu.:-1.11862 1st Qu.:-0.6660
## Median :-0.11862 Median :-0.1395
## Mean :-0.00022 Mean : 0.0000
## 3rd Qu.: 1.30995 3rd Qu.: 0.5493
## Max. : 3.80995 Max. : 5.9795
## NA's :42 NA's :462.5 Standardising Data
If you haven’t done this before creating the unmarkedFrame object the syntax for subsetting changes slightly since an unmarkedFrame is an S4 class. The first $ subset is replaced with an @
umf@siteCovs$elev <- scale(umf@siteCovs$elev)
umf@siteCovs$forest <- scale(umf@siteCovs$forest)
umf@siteCovs$length <- scale(umf@siteCovs$length)
umf@obsCovs$date <- scale(umf@obsCovs$date)
umf@obsCovs$ivel <- scale(umf@obsCovs$ivel)2.6 Missing Data Considerations
We have two types of data in these models: our observation data and our covariate data. These models account for missing data in the two groups differently.
- You are allowed to have missing records in your observation data. In practice this could result from varying numbers of repeat visits to a site or the loss of data. As long as a site has at least one observation it can be used in the model. Any site that has zero observations will be removed.
- You cannot have missing covariate data. If a site is missing the data for a site-level covariate it will be removed before model fitting. If a site is missing observation-level covariate data it will only be removed if it is missing for all observations, but the observation with missing data is removed. This is very important to consider when performing model selection so we will touch on this again later.
2.7 Fitting a Model
Fitting a model with unmarked uses a similar syntax to fitting a linear model with the lm() function. The main difference is that the way a formula is defined varies. In the case of occu() it requires a double right-hand side formula for detection and occupancy covariates in that order. Essentially ~ detection formula ~ occupancy formula.
##
## Call:
## occu(formula = ~1 ~ 1, data = umf)
##
## Occupancy:
## Estimate SE z P(>|z|)
## -0.665 0.139 -4.77 1.82e-06
##
## Detection:
## Estimate SE z P(>|z|)
## 1.32 0.174 7.61 2.82e-14
##
## AIC: 528.987unmarked estimates are on the link-scale (logit for occu() since it uses a logit-link), and the backTransform() function coverts them back to the original scale. You need to specify a type of state or det for occupancy or detection covariates. If you have fit a model with covariates then you need to specify values for them (i.e. what is the probability of occupancy when CovA = X and CovB = Y)
## Backtransformed linear combination(s) of Occupancy estimate(s)
##
## Estimate SE LinComb (Intercept)
## 0.34 0.0313 -0.665 1
##
## Transformation: logistic## Backtransformed linear combination(s) of Detection estimate(s)
##
## Estimate SE LinComb (Intercept)
## 0.79 0.0289 1.32 1
##
## Transformation: logisticAlternatively, you can define an antilogit function:
## [1] 0.3396173## [1] 0.7891817Now, to add some covariates. First, assuming constant detection:
##
## Call:
## occu(formula = ~1 ~ forest + elev + length, data = umf)
##
## Occupancy:
## Estimate SE z P(>|z|)
## (Intercept) -1.05 0.195 -5.373 7.74e-08
## forest 1.21 0.211 5.734 9.79e-09
## elev 1.39 0.229 6.070 1.28e-09
## length 0.14 0.179 0.784 4.33e-01
##
## Detection:
## Estimate SE z P(>|z|)
## 1.32 0.175 7.53 5.13e-14
##
## AIC: 453.6822And some more:
##
## Call:
## occu(formula = ~date + ivel + forest ~ forest + elev + length,
## data = umf)
##
## Occupancy:
## Estimate SE z P(>|z|)
## (Intercept) -0.154 0.320 -0.482 6.30e-01
## forest 0.339 0.305 1.110 2.67e-01
## elev 2.328 0.416 5.591 2.26e-08
## length 0.472 0.242 1.950 5.12e-02
##
## Detection:
## Estimate SE z P(>|z|)
## (Intercept) 0.2350 0.197 1.193 2.33e-01
## date 0.0994 0.176 0.566 5.71e-01
## ivel 0.5050 0.218 2.319 2.04e-02
## forest 1.5522 0.221 7.019 2.24e-12
##
## AIC: 433.60142.8 Model Selection
2.8.1 unmarked
The is an in-built model selection method using fitList() and modSel(). You can name the models whatever you like, but the convention used here is common online (even if awkward to type out, especially for large numbers of covariates).
fit <- fitList('psi(.)p(.)' = fm,
'psi(forest + elev + length)p(.)' = fm1,
'psi(forest + elev + length)p(date + ivel + forest)' = fm2)
modSel(fit)## nPars
## psi(forest + elev + length)p(date + ivel + forest) 8
## psi(forest + elev + length)p(.) 5
## psi(.)p(.) 2
## AIC
## psi(forest + elev + length)p(date + ivel + forest) 433.60
## psi(forest + elev + length)p(.) 453.68
## psi(.)p(.) 528.99
## delta
## psi(forest + elev + length)p(date + ivel + forest) 0.00
## psi(forest + elev + length)p(.) 20.08
## psi(.)p(.) 95.39
## AICwt
## psi(forest + elev + length)p(date + ivel + forest) 1.0e+00
## psi(forest + elev + length)p(.) 4.4e-05
## psi(.)p(.) 1.9e-21
## cumltvWt
## psi(forest + elev + length)p(date + ivel + forest) 1.00
## psi(forest + elev + length)p(.) 1.00
## psi(.)p(.) 1.002.8.2 MuMIn
All unmarked models are compatible with the functions in the MuMIn package. For example, the dredge() and pdredge() functions perform dredge-based model selection that takes a ‘full’ or ‘global’ model and fits every possible combination of covariates and ranks them by some information criteria (AIC, BIC, etc.).
full <- occu(formula = ~ date + ivel + forest
~ forest + elev + length,
data = umf)
modelList <- dredge(full,
rank = "AIC")## Fixed terms are "p(Int)" and "psi(Int)"| p(Int) | psi(Int) | p(date) | p(forest) | p(ivel) | psi(elev) | psi(forest) | psi(length) | df | logLik | AIC | delta | weight | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 0.2717989 | 0.1371948 | NA | 1.5844055 | 0.3213761 | 2.5016604 | NA | NA | 5 | -210.4061 | 430.8122 | 0.0000000 | 2.002183e-01 |
| 47 | 0.2270574 | 0.0809094 | NA | 1.5670653 | 0.5224874 | 2.4738438 | NA | 0.5137398 | 6 | -209.7004 | 431.4009 | 0.5886862 | 1.491668e-01 |
| 16 | 0.2030971 | 0.1593233 | 0.1664722 | 1.6612138 | 0.2948331 | 2.4769948 | NA | NA | 6 | -209.9298 | 431.8596 | 1.0474156 | 1.185933e-01 |
| 63 | 0.2774926 | -0.1777917 | NA | 1.5045006 | 0.5309495 | 2.3354733 | 0.3610538 | 0.4745035 | 7 | -208.9608 | 431.9217 | 1.1095044 | 1.149682e-01 |
| 31 | 0.3012686 | -0.0227557 | NA | 1.5541616 | 0.3184063 | 2.3982256 | 0.2026555 | NA | 6 | -210.2111 | 432.4221 | 1.6099352 | 8.951808e-02 |
| 48 | 0.1788329 | 0.0982908 | 0.1319485 | 1.6263881 | 0.4746579 | 2.4564416 | NA | 0.4893203 | 7 | -209.4140 | 432.8279 | 2.0157310 | 7.307912e-02 |
| 64 | 0.2350160 | -0.1543441 | 0.0994379 | 1.5521741 | 0.5050283 | 2.3283553 | 0.3389721 | 0.4716440 | 8 | -208.8007 | 433.6014 | 2.7892145 | 4.964020e-02 |
| 32 | 0.2306313 | 0.0403105 | 0.1517278 | 1.6329933 | 0.2928017 | 2.4085725 | 0.1462925 | NA | 7 | -209.8344 | 433.6688 | 2.8566108 | 4.799529e-02 |
| 11 | 0.4466097 | 0.0915580 | NA | 1.4744212 | NA | 2.5637670 | NA | NA | 4 | -213.0908 | 434.1815 | 3.3693343 | 3.714173e-02 |
| 12 | 0.3324200 | 0.1241412 | 0.2308726 | 1.5993875 | NA | 2.5190296 | NA | NA | 5 | -212.1499 | 434.2997 | 3.4875171 | 3.501056e-02 |
| 27 | 0.4793926 | -0.1087246 | NA | 1.4412648 | NA | 2.4230182 | 0.2761855 | NA | 5 | -212.6709 | 435.3418 | 4.5295744 | 2.079310e-02 |
| 28 | 0.3685889 | -0.0388545 | 0.2095462 | 1.5629547 | NA | 2.4202416 | 0.2118323 | NA | 6 | -211.9236 | 435.8471 | 5.0349557 | 1.615017e-02 |
| 43 | 0.4525069 | 0.0889259 | NA | 1.4713281 | NA | 2.6091039 | NA | 0.1634937 | 5 | -212.9256 | 435.8513 | 5.0391041 | 1.611670e-02 |
| 44 | 0.3385965 | 0.1197464 | 0.2282498 | 1.5945437 | NA | 2.5610702 | NA | 0.1531902 | 6 | -212.0063 | 436.0126 | 5.2004500 | 1.486758e-02 |
| 59 | 0.4884183 | -0.1255346 | NA | 1.4335076 | NA | 2.4685178 | 0.2959053 | 0.1894972 | 6 | -212.4510 | 436.9020 | 6.0898420 | 9.530405e-03 |
| 60 | 0.3787542 | -0.0597685 | 0.2051483 | 1.5517870 | NA | 2.4593252 | 0.2348551 | 0.1782155 | 7 | -211.7321 | 437.4643 | 6.6520623 | 7.194933e-03 |
| 25 | 1.3157034 | -1.0496155 | NA | NA | NA | 1.3475082 | 1.2191191 | NA | 4 | -222.1556 | 452.3113 | 21.4990613 | 4.295778e-06 |
| 26 | 1.2844601 | -1.0522406 | 0.1718313 | NA | NA | 1.3354226 | 1.2242119 | NA | 5 | -221.6936 | 453.3873 | 22.5751001 | 2.508320e-06 |
| 29 | 1.2445959 | -1.0455354 | NA | NA | 0.1848135 | 1.3455297 | 1.2218243 | NA | 5 | -221.7782 | 453.5564 | 22.7441771 | 2.304987e-06 |
| 57 | 1.3157574 | -1.0456432 | NA | NA | NA | 1.3892074 | 1.2105149 | 0.1401486 | 5 | -221.8411 | 453.6822 | 22.8700249 | 2.164417e-06 |
| 58 | 1.2839466 | -1.0482557 | 0.1728783 | NA | NA | 1.3776263 | 1.2154396 | 0.1409657 | 6 | -221.3706 | 454.7412 | 23.9290310 | 1.274620e-06 |
| 61 | 1.2377919 | -1.0394241 | NA | NA | 0.1965896 | 1.3918236 | 1.2121429 | 0.1521085 | 6 | -221.4105 | 454.8210 | 24.0088570 | 1.224748e-06 |
| 30 | 1.2355377 | -1.0486673 | 0.1406246 | NA | 0.1412025 | 1.3357680 | 1.2252015 | NA | 6 | -221.4910 | 454.9820 | 24.1698020 | 1.130051e-06 |
| 62 | 1.2295468 | -1.0429011 | 0.1385517 | NA | 0.1531505 | 1.3811610 | 1.2158195 | 0.1494977 | 7 | -221.1312 | 456.2624 | 25.4501902 | 5.957515e-07 |
| 24 | 0.2291546 | -0.3662072 | 0.3609984 | 1.2961532 | 0.7285032 | NA | 0.4142468 | NA | 6 | -240.1228 | 492.2456 | 61.4333947 | 9.149799e-15 |
| 8 | -0.0281270 | -0.0475340 | 0.3844998 | 1.6052211 | 0.8287601 | NA | NA | NA | 5 | -241.6817 | 493.3634 | 62.5512227 | 5.232127e-15 |
| 56 | 0.2664051 | -0.4019090 | 0.3551751 | 1.2641240 | 0.6982385 | NA | 0.4515160 | -0.0753654 | 7 | -240.0298 | 494.0595 | 63.2473534 | 3.694158e-15 |
| 23 | 0.3842317 | -0.4417472 | NA | 1.1495425 | 0.7932479 | NA | 0.4703237 | NA | 5 | -242.2418 | 494.4836 | 63.6714068 | 2.988363e-15 |
| 40 | -0.0341475 | -0.0408255 | 0.3867366 | 1.6029884 | 0.8404482 | NA | NA | 0.0485674 | 6 | -241.6422 | 495.2844 | 64.4722034 | 2.002362e-15 |
| 55 | 0.4367117 | -0.4864980 | NA | 1.1029636 | 0.7444892 | NA | 0.5199886 | -0.1109263 | 6 | -242.0311 | 496.0622 | 65.2500407 | 1.357180e-15 |
| 9 | 1.2440471 | -0.7238769 | NA | NA | NA | 0.9071172 | NA | NA | 3 | -245.2019 | 496.4038 | 65.5916172 | 1.144103e-15 |
| 41 | 1.2392010 | -0.7231367 | NA | NA | NA | 0.9940344 | NA | 0.2375585 | 4 | -244.2499 | 496.4999 | 65.6876733 | 1.090452e-15 |
| 7 | 0.0817147 | -0.0911363 | NA | 1.5057697 | 0.9449782 | NA | NA | NA | 4 | -244.3417 | 496.6834 | 65.8711944 | 9.948454e-16 |
| 45 | 1.1346845 | -0.7114837 | NA | NA | 0.2496379 | 1.0088969 | NA | 0.2674159 | 5 | -243.6462 | 497.2924 | 66.4802152 | 7.336829e-16 |
| 13 | 1.1696647 | -0.7199777 | NA | NA | 0.2033234 | 0.9001801 | NA | NA | 4 | -244.8389 | 497.6777 | 66.8655382 | 6.051132e-16 |
| 10 | 1.2380311 | -0.7311123 | 0.0959000 | NA | NA | 0.8852939 | NA | NA | 4 | -245.0790 | 498.1580 | 67.3458410 | 4.759268e-16 |
| 42 | 1.2326285 | -0.7304323 | 0.0995715 | NA | NA | 0.9709839 | NA | 0.2350621 | 5 | -244.1140 | 498.2280 | 67.4157894 | 4.595693e-16 |
| 20 | 0.8339574 | -0.6808379 | 0.4001558 | 0.7679528 | NA | NA | 0.6833542 | NA | 5 | -244.1575 | 498.3149 | 67.5027332 | 4.400190e-16 |
| 39 | 0.0785087 | -0.0875094 | NA | 1.5047510 | 0.9518323 | NA | NA | 0.0269958 | 5 | -244.3292 | 498.6584 | 67.8461669 | 3.705918e-16 |
| 52 | 0.8389858 | -0.6870414 | 0.3882027 | 0.7653299 | NA | NA | 0.7103056 | -0.1974959 | 6 | -243.3853 | 498.7706 | 67.9584442 | 3.503605e-16 |
| 46 | 1.1380968 | -0.7150126 | 0.0360656 | NA | 0.2357348 | 0.9982608 | NA | 0.2636282 | 6 | -243.6305 | 499.2611 | 68.4488704 | 2.741703e-16 |
| 14 | 1.1728537 | -0.7244779 | 0.0552900 | NA | 0.1846012 | 0.8879189 | NA | NA | 5 | -244.8010 | 499.6021 | 68.7899069 | 2.311880e-16 |
| 19 | 1.0712302 | -0.7239623 | NA | 0.5284574 | NA | NA | 0.7187187 | NA | 4 | -246.3815 | 500.7631 | 69.9508739 | 1.293792e-16 |
| 51 | 1.0614332 | -0.7253322 | NA | 0.5431030 | NA | NA | 0.7427912 | -0.2105270 | 5 | -245.5000 | 500.9999 | 70.1877339 | 1.149294e-16 |
| 17 | 1.3384933 | -0.7556707 | NA | NA | NA | NA | 0.7644613 | NA | 3 | -248.7650 | 503.5299 | 72.7177422 | 3.243745e-17 |
| 18 | 1.2778608 | -0.7514852 | 0.2275269 | NA | NA | NA | 0.7692585 | NA | 4 | -247.9271 | 503.8542 | 73.0419657 | 2.758306e-17 |
| 49 | 1.3369779 | -0.7601678 | NA | NA | NA | NA | 0.7921068 | -0.1987465 | 4 | -247.9441 | 503.8883 | 73.0760790 | 2.711657e-17 |
| 50 | 1.2780446 | -0.7569066 | 0.2245248 | NA | NA | NA | 0.7972470 | -0.1958420 | 5 | -247.1296 | 504.2592 | 73.4470266 | 2.252602e-17 |
| 21 | 1.2426829 | -0.7478339 | NA | NA | 0.2229338 | NA | 0.7667187 | NA | 4 | -248.2337 | 504.4674 | 73.6551805 | 2.029946e-17 |
| 53 | 1.2506170 | -0.7540493 | NA | NA | 0.2082875 | NA | 0.7933912 | -0.1905279 | 5 | -247.4794 | 504.9587 | 74.1465435 | 1.587766e-17 |
| 22 | 1.2103185 | -0.7449714 | 0.1969607 | NA | 0.1730673 | NA | 0.7710529 | NA | 5 | -247.6368 | 505.2737 | 74.4614681 | 1.356442e-17 |
| 54 | 1.2194474 | -0.7518920 | 0.1961805 | NA | 0.1566134 | NA | 0.7977083 | -0.1897549 | 6 | -246.8906 | 505.7812 | 74.9690300 | 1.052412e-17 |
| 4 | 0.2541686 | -0.2009794 | 0.5594061 | 1.5617529 | NA | NA | NA | NA | 4 | -249.8103 | 507.6206 | 76.8084088 | 4.195364e-18 |
| 36 | 0.2357155 | -0.1708860 | 0.5396890 | 1.5978246 | NA | NA | NA | -0.2291602 | 5 | -249.1665 | 508.3329 | 77.5207357 | 2.938257e-18 |
| 3 | 0.5617480 | -0.3685693 | NA | 1.2354923 | NA | NA | NA | NA | 3 | -255.5722 | 517.1444 | 86.3322160 | 3.586751e-20 |
| 35 | 0.5167888 | -0.3205897 | NA | 1.3043660 | NA | NA | NA | -0.2659546 | 4 | -254.6159 | 517.2317 | 86.4195251 | 3.433541e-20 |
| 1 | 1.3249166 | -0.6650338 | NA | NA | NA | NA | NA | NA | 2 | -262.4935 | 528.9870 | 98.1747779 | 9.618815e-23 |
| 2 | 1.2749717 | -0.6639879 | 0.2096239 | NA | NA | NA | NA | NA | 3 | -261.7925 | 529.5850 | 98.7728449 | 7.132684e-23 |
| 5 | 1.2234967 | -0.6563222 | NA | NA | 0.2367346 | NA | NA | NA | 3 | -261.9381 | 529.8763 | 99.0640771 | 6.166130e-23 |
| 33 | 1.3236477 | -0.6656290 | NA | NA | NA | NA | NA | -0.0878263 | 3 | -262.2988 | 530.5975 | 99.7853108 | 4.299310e-23 |
| 6 | 1.2000292 | -0.6562906 | 0.1768538 | NA | 0.1903936 | NA | NA | NA | 4 | -261.4647 | 530.9294 | 100.1171656 | 3.641979e-23 |
| 34 | 1.2752897 | -0.6649410 | 0.2066496 | NA | NA | NA | NA | -0.0821538 | 4 | -261.6203 | 531.2406 | 100.4284182 | 3.117093e-23 |
| 37 | 1.2280744 | -0.6582929 | NA | NA | 0.2282182 | NA | NA | -0.0761202 | 4 | -261.7907 | 531.5814 | 100.7691800 | 2.628780e-23 |
| 38 | 1.2051423 | -0.6582980 | 0.1756097 | NA | 0.1817536 | NA | NA | -0.0738349 | 5 | -261.3252 | 532.6504 | 101.8381625 | 1.540379e-23 |
2.8.3 Missing Data
As mentioned previously, missing data can have serious impacts on model selection. This is because information criterion like AIC are not comparable between models fit to different datasets. If you fit Model A with covariates 1 & 2 with 100 sites, and Model B with covariates 1, 2, & 3 with 90 sites (because covariate 3 is missing data) then you cannot compare AIC. This rule holds no matter how minuscule the difference is (e.g. 10,000 vs 9,999).
To account for this you need to manually remove the data for all sites/observations that are missing data for any covariates that could potentially be included in your model. In the previous example, both Model A and Model B should be fit to the 90 sites not missing data for covariate 3.
2.9 Proportion of Area Occupied
Imperfect detection can bias our occupancy estimates. So how do we calculate the actual proportion of area occupied by our target species while accounting for imperfect detection?
2.9.1 Assuming perfect detection
If we naively assumed perfect detection the proportion of area occupied by a species is simply the proportion of occupied sites: number of sites where the species was observed divided by the total number of sites.
## [1] 0.33333332.9.2 Accounting for Imperfect Detection
In reality we wouldn’t assume perfect detection, so we need to account for it otherwise we will end up with a biased underestimate of the proportion of area occupied. unmarked utilises empirical Bayes methods to estimate the posterior distributions of the random variables (here, latent occurrence state, \(z_i\)).
\[Pr(z_i = 1 | y_{ij}, \psi_i, p_{ij})\]
AICbest <- occu(formula = ~ forest + ivel
~ elev,
data = umf)
re <- ranef(AICbest)
EBUP <- bup(re, stat="mean")
CI <- confint(re, level=0.9)
rbind(PAO = c(Estimate = sum(EBUP), colSums(CI)) / 237)## Estimate 5% 95%
## PAO 0.4858397 0.4219409 0.5949367A difference in the proportion of area occupied of over 15%! Depending on how hard a species can be to detect with a given survey method this difference can potentially be much higher!
2.10 Prediction
Lets load in a bigger dataset for this one. This is a dataset for ink card surveys for the giant gecko, Cyrtodactylus sadleiri, on Christmas Island.
gg <- read.csv("Data/GG.csv")
umfGG <- unmarkedFrameOccu(y = gg[ ,2:11],
siteCovs = gg[ ,52:55],
obsCovs = list(BHG = gg[ ,12:21],
Rat = gg[ ,22:31],
Height = gg[ ,32:41],
Orientation = gg[ ,42:51]))Now lets fit a model with a larger amount of covariates. We can also alter some additional model parameters. method sets to optimisation method used by optim under the hood to estimate the maximum likelihood, control here sets the maximum number of iterations allowed for the model to converge (the default value is rather small so this is useful for more complex models), and engine gives you the choice to run the optimisation in native R code or fast C++.
occuGG <- occu(formula = ~ BHG + Rat + Height + Orientation +
DistRoad.S + Slope.S + I(Slope.S^2)
~ YCASC + CHM.S + DistRoad.S,
data = umfGG,
method = "BFGS",
control = list(maxit = 10000),
engine = "C")## Warning: Some observations have been discarded
## because corresponding covariates were missing.## Warning: 100 sites have been discarded because of
## missing data.Now we load in the data we want to predict occupancy for. This dataset needs to have two columns for longitude and latitude values for each site/pixel, and a column for each covariate (each row is one site/pixel). Categorical variables need to have a single level specified per site. This is straight forward for something like vegetation class, but for methodological variables like survey method(ink cards, spotlighting, etc.) you need to select one a priori.
And predict.
## doing row 1000 of 27593
## doing row 2000 of 27593
## doing row 3000 of 27593
## doing row 4000 of 27593
## doing row 5000 of 27593
## doing row 6000 of 27593
## doing row 7000 of 27593
## doing row 8000 of 27593
## doing row 9000 of 27593
## doing row 10000 of 27593
## doing row 11000 of 27593
## doing row 12000 of 27593
## doing row 13000 of 27593
## doing row 14000 of 27593
## doing row 15000 of 27593
## doing row 16000 of 27593
## doing row 17000 of 27593
## doing row 18000 of 27593
## doing row 19000 of 27593
## doing row 20000 of 27593
## doing row 21000 of 27593
## doing row 22000 of 27593
## doing row 23000 of 27593
## doing row 24000 of 27593
## doing row 25000 of 27593
## doing row 26000 of 27593
## doing row 27000 of 27593levelplot(Predicted ~ ggPred$x + ggPred$y,
data = occuPred,
col.regions = rev(terrain.colors(100)),
at = seq(0,1,length.out=101))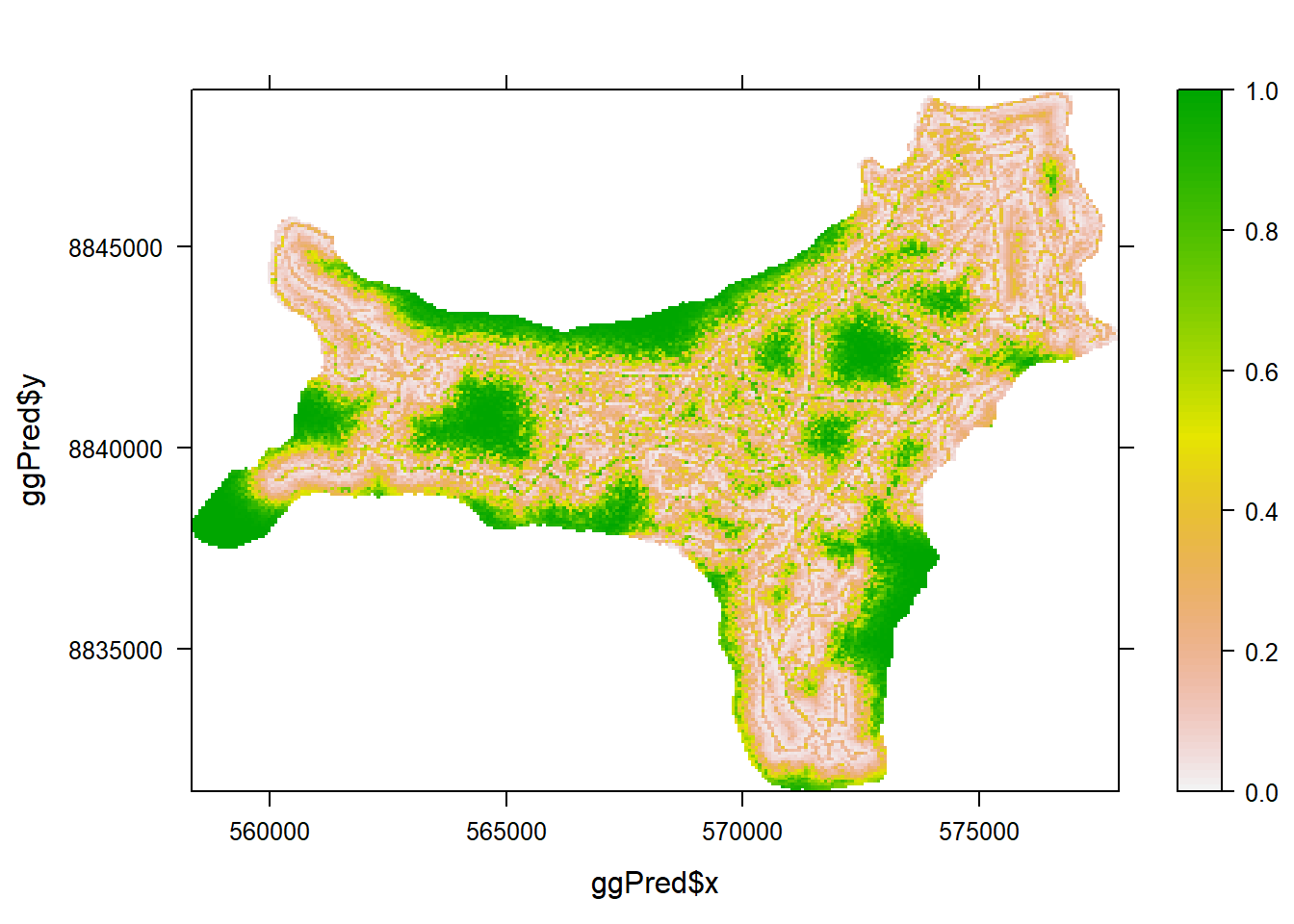
2.11 Model Evaluation
A vital step in modelling is evaluating the accuracy of model predictions. Model evaluation usually involves testing how well our model can predict data not used in the model fitting process. Does our model predict high probability of occupancy for sites with observed presences in our hold-out data? Does it predict low probability of ocurrence for the sites with observed absences? The Area Under the Receiver Operating Curve (AUC) is a popular metric for this evaluation.
The problem with this type of model evaluation for models accounting for imperfect detection is that we don’t know the true occupancy state of a site. Is the species actually absent from the site in our hold-out data, or did we just fail to detect it? As such this type of model evaluation is not appropriate for our modelling method.
Instead we can use several goodness-of-fit tests to perform model evaluation. Here we use three tests: sum of squared errors, Pearson’s Chi-squared, and Freeman-Tukey Chi-squared. We create a new function called fitstats (where model.name should be replaced with your own), and then use it when fitting our model to bootsrapped samples of our data to assess model fit. Even though these are called goodness-of-fit tests they are looking for significant lack-of-fit evidence (which sounds too pessimistic), so a p-value of <0.05 is actually a poorly fit model. A “good” model should return a value >> 0.05 for each test. Yet the issue of imperfect detection can still impact these tests. It is possible to fail one test but pass the others on the basis of poor prediction at a small subset of sites (even one). For example, an observed presence at a really low probability of detection site can dramatically inflate the test statistic. In these scenarios it is important to assess the input of each site to the test statistic to determine if the “lack-of-fit” can be disregarded as over-penalising the mismatch in a very low proportion of sites.
fitstats <- function(model.name,
method = "nonparboot") {
observed <- getY(model.name@data)
expected <- fitted(model.name)
resids <- residuals(model.name,
method = "nonparboot")
sse <- sum(resids^2,
na.rm = TRUE)
chisq <- sum((observed - expected)^2 / expected,
na.rm = TRUE)
freeTuke <- sum((sqrt(observed) - sqrt(expected))^2,
na.rm = TRUE)
out <- c(SSE = sse,
Chisq = chisq,
freemanTukey = freeTuke)
return(out)
}
pb <- parboot(model.name,
fitstats,
nsim = 1000,
report = TRUE,
method = "nonparboot")
pb
par(mfrow = c(3,1))
plot(pb,
main = "",
xlab = c("SSE", "Chisq", "FT"))2.12 Common Errors
- Error: Hessian is singular. Try providing starting values or using fewer covariates. (The bane of my Masters existence)
- This could come about for a variety of reasons.
- For models with a very low naive occupancy rate, especially in small datasets, you might need to specify starting values for the maximum likelihood estimation (see
?occu) - You have complete separation in one or more covariates. This means the model doesn’t have the necessary data to calculate parameter estimates. For categorical variables this is commonly a factor level that has exclusively presence or absence observations (or potentially when the prevalence is near 0 or 1 in very big datasets), or there are factor levels in two or more categorical variables that never co-occur. Consider merging factor levels if possible! For continuous variables this is where the data is split into two (or more) groups at either end of a sampling range. For example, you are including elevation as a covariate but have presence records only at 0-20m, and absences only at 80-100m. The model then can’t estimate the slope of the relationship.
- You have somehow got a continuous variable set to factor
- You have too many covariates in the model for your data. There is no set rule for this and different people will give different answers. Every 10 sites allows one covariate, every 10 presences allows one covariate, every 10 allows one covariate in each formula, each level in a categorical variable is treated as a separate binary dummy variable and it does/does not count as a separate covariate for this count etc..
- Something about factor levels and prediction.
- If you have a model with a categorical covariate with 5 factor levels, the data being predicted to must have 5 levels. Each site being predicted to can have the same level, but the data must be allowed to have other levels. This is an
Rerror and not a statistical error. This is commonly caused by methodological observation-level covariates. For example, your data comes from three different survey methods (e.g. spotlighting, trapping, and drones), you fit the model and find out that spotlighting has the best probability of detecting your target species, and then predict to only spotlighting as a survey method. A really easy mistake to make! - A dodgy fix: Set a random cell on the border of the area you are predicting to to the missing factor level
- A proper fix: Use the
levels()function to set additional levels to your prediction data
- If you have a model with a categorical covariate with 5 factor levels, the data being predicted to must have 5 levels. Each site being predicted to can have the same level, but the data must be allowed to have other levels. This is an
- Your fitted model has unrealistic parameter estimates.
occu()is on the logit-scale, so anything outside [-5, 5] is when you need to start checking. When this is back transformed to the probability scale it is roughly [0, 1]. There is leeway if you are considering polynomial terms. If you start getting parameter estimates of +30, -90, or +462.10 (David’s high score) then you definitely have an issue! This is equally likely to happen to categorical or continuous data.- This could be one of two issues:
- Your model has not converged on the maximum likelihood. Use the
controlargument when fitting the model to increase the number of iterations in the optimisation process. The default is rather small at 100 or 500 depending on your optimisation method. Setting it high (i.e. 10,000) is fine, as it is a maximum, and the model will stop if it converges earlier. - Your model has converged on a local maximum. Likelihoods can have multiple peaks, but only one maximum, and your model is stuck on the wrong one. To fix this use the
startsargument to provide starting values for parameters to start the optimisation in a different area.
- Your model has not converged on the maximum likelihood. Use the
- Model selection with
dredge()is too much for my computer to handle. + See David (for Spartan) or Casey/Nick (for Boab)
2.13 Additional resources
- Overview of unmarked vignette by Ian Fiske and Richard Chandler. The official intro guide that I copied some of the early examples from.
- unmarked: An R Package for Fitting Hierarchical Models of Wildlife Occurrence and Abundance. The paper about the package
- The
unmarkedwebsite. - The
unmarkedgoogle group/forum. This is frequented by most of the package creators/contributors and other occupancy modellers. More often than not the answer to your problems has already been solved here.